Data Science : Making sense of High Dimensional Data using TSNE — Part II
Understanding the intuition and mathematics behind mapping high dimension spaces to low dimension spaces using TSNE

Table of Contents —
1) Continuation of Previous Blog (Part 1)
2) Dimensionality Reduction Technique : TSNE
- TSNE Intuition
- TSNE Mathematics and algorithm
- TSNE Application on the dataset
- TSNE final notes
1) Continuation of Previous Blog (Part 1)
Refer to my previous blog post for understanding the problem statement —
You can continue reading after that post from here below. This is the Part -2 of Dimensionality Reduction technique. In this article, we will understand in depth of the TSNE technique and how it compresses data into a lower dimension space by preserving local neighbourhood embeddings.
2) Dimensionality Reduction Technique : TSNE
TSNE Intuition —
TSNE stands for t-distributed stochastic neighbourhood embedding. Intuitively, what it does is when mapping the dataset from 30 D to 2 D, it preserves the local neighbourhood distances which basically in this context would mean that patients who are very similar (neighbours) in their 30 characteristic space would also be neighbours in the 2D space. And then the mappings in the 2D space is learnt in a way so as to preserve (minimize) the local neighbourhood distances as much as possible across all the points.
TSNE Mathematics and Algorithm —
The exact Mathematics of TSNE are quite Math intense to a large extent. You can read the original TSNE research paper here — https://lvdmaaten.github.io/publications/papers/JMLR_2008.pdf
I’ll try to explain in brief what Stochastic Neighbourhood Embedding (SNE) is in short to get the gist of the algorithm.
(Step 1) : Calculate the Pairwise affinities between all the points in the higher dimension space.
The affinity between 2 points xi and xj in the high dimension space is given by —

Let us understand this above equation in English.
This basically represents the Conditional Probability of selecting point ‘j’ as a neighbour of point ‘i’. So the affinity of point ‘j’ given you are at point ‘i’ is nothing but the sum normalized Gaussian Kernel (Radial Basis Function Kernel) also commonly known as (RBF Kernel) values across your neighbourhood points if you had to pick a point when you are standing at the point ‘i’ (Mean) and assume you take a variance of sigma_i. This value sigma_i is a value that you are indirectly telling how many neighbours you want to consider in the probability calculation of the above equation. In other words, this is roughly akin to the Number of Neighbours you want to consider.
If you are interested in knowing more about RBF Kernel (Gaussian Kernel) — you can check the link — https://en.wikipedia.org/wiki/Radial_basis_function_kernel
Calculating sigma_i (using Perplexity hyperparameter) :
As per the research paper above, sigma / (std. deviation) for a point ‘i’ differs for every point since the density of points varies across the dataset. Now, by assuming a Gaussian centered at point ‘i’ with sigma_i as the standard deviation, the PDF is obtained across all points.
The perplexity is defined as the 2nd exponent of the Shannon entropy of the PDF distribution as generated above.


So, when we specify the Perplexity as the hyperparameter, indirectly we can think that we are defining the Shannon Entropy that we want to get across all points. So from the Entropy we can back calculate the sigma_i value given we define our perplexity.
Now, while you select the optimum perplexity value, you might want to remember that —
-> Why do we use Shannon Entropy and why not just the simple Standard deviation ? One of the reasons for using Shannon Entropy in the research paper would be that Entropy is more robust to Multi-Modal distributions than a simple standard deviation which do not capture Modality of the dataset. So, Hypothetically, if we were to change the distribution from Gaussian to other distribution assumptions, it’s better to use Shannon Entropy.
-> Now, if we are defining higher values of perplexity, that means we get higher values of Entropy which means the Gaussian distribution will eventually get more flattened. This means the farther distant points also get a relatively significant affinity as compared to the near points which will have the highest affinity. We can intuitively think of this as ‘K’ from K-Nearest Neighbours search where if you increase the ‘K’ value, we get a larger neighbourhood of points to consider our sampling from.
(Step 2) : Sample an initial solution in the low dimension space across all points from an Isotropic Gaussian distribution
Let’s say you are projecting into a 2 DIM low space. This basically means you are sampling each of the points from a bivariate normal distribution. So you sample each point from a joint normal distribution with mean vector as all zeros (low D space) and the covariance matrix as a scalar multiplier of the Identity Matrix (Isotropic Gaussian case).


If you want to know more about Bivariate Isotropic Gausssian distribution, you can refer to this link — https://math.stackexchange.com/questions/1991961/gaussian-distribution-is-isotropic#:~:text=TLDR%3A%20An%20isotropic%20gaussian%20is,%CE%A3%20is%20the%20covariance%20matrix.
np.random.multivariate_normal(mean =[0,0],cov = np.identity(2)*1e-4)(Step 3) : Use Gradient Descent algorithm to learn the best Low dimension embeddings that minimizes the overall KL divergence across all the data points
Upto Step 2, we have obtained a feature mapping in a 2D space of each point but this was obtained from sampling.
But, how do we ensure that the mappings obtained in 2D low space are locally close to that of the high D space ? We get this by learning the low dimension mappings by minimizing a cost function. This cost function should be indicative of the closeness of the high D and low D mappings. A common metric for this is to use the Sum of KL divergences across all points. The metric for cost function is the KL divergence across all points —

Why KL divergence ?
Because KL divergence compares similarity of 2 distributions. The more similar 2 distributions are, the lower their KL divergence will be. Basically, KL divergence (p,q) between 2 distributions parametrized by (p,q) respectively is the distance between the 2 log-probabilities with a weightage factor of p_i.
How to get a symmetric measure of the KL divergence ?
You might have understood that the conditional probability p(i | j) will not be equal to p(j | i). The way they implement a symmetric measure of KL divergence is to take the joint probability distribution instead of the conditional probability distribution. The joint probability distribution is estimated as —

How to get the lower dimension affinities (q_ij) ?
This is obtained in the same way like we calculated for p_ij , The low dimension affinity between 2 points is —

The only difference that you will notice is that there is no sigma term — that’s because it is just taken as 1/sqrt(2). So essentially the denominator in the exponent term becomes 1 to keep the math simple. Plus, in a 2 D space, we will be much less worried about wether sigma_i (which is a measure of variability while defining our affinities) would vary a lot across each of the data points as compared to a high dimensional space.
Now, that we have a definite value of the Cost Function between high dimension maps and low dimension maps, all we need to do is keep readjusting the low dimension maps in a way so as to minimize the cost function above over multiple iterations using Gradient Descent.
The first equation below is the Derivative of the Cost Function.
The second equation is the update term of the popular Gradient Descent algorithm along with the momentum term (marked in Red).


Why use Momentum term in update equation ?
As you can see from the equation above that Momentum is nothing but the exponential weighing of the difference of the previous gradients. So instead of taking the gradient only of the current iteration, we also take the gradients obtained in the previous iterations where the weights of these previous gradients are decayed exponentially. This has the effect of reducing oscillations as the local minima / maxima is being reached.
TSNE application on the dataset —
We need to try running TSNE with multiple values of perplexity. On running the TSNE on the above dataset with perplexities of —
perplexity = [3,5,9,15,25,50]On running TSNE with each of the perplexity values above, we get the following plots —




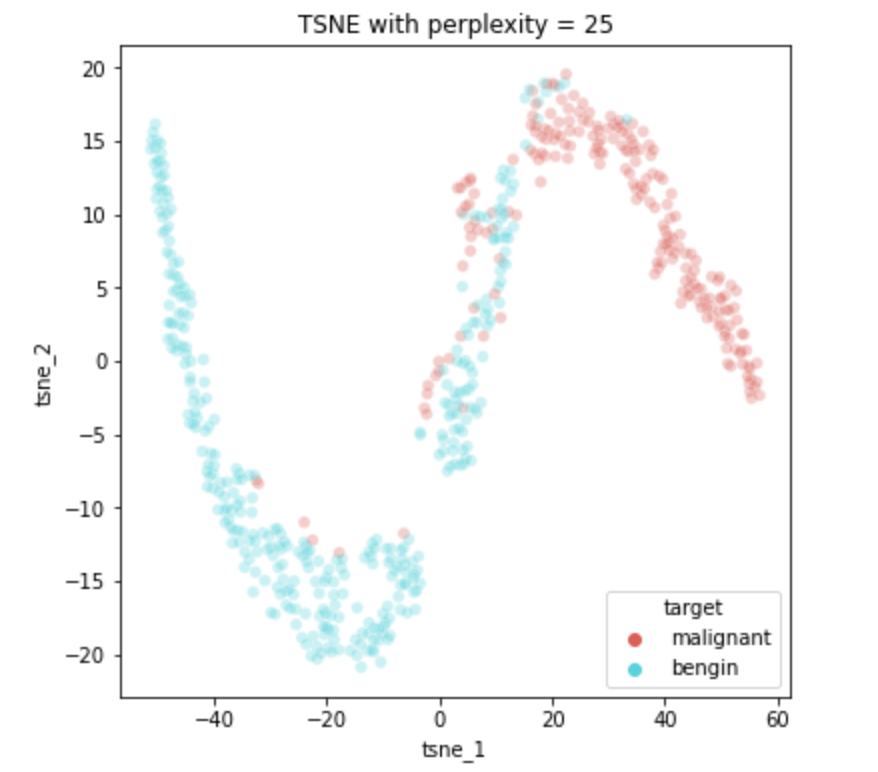

We can clearly see that for higher ranged values of perplexity (25, 50), we get a very clear seperation of the malignant and bengin patients in a 2D space which has been mapped from a 30D space by preserving local neighbourhoods.
TSNE final Notes —
- It is recommended to run TSNE with multiple values of perplexity and check what value gives a good representation in a lower dimension space. This can be treated as a hyper parameter for your problem statement.
- TSNE gives different outputs on every run because it is a stochastic algorithm where each output of TSNE will be different from the previous one since every output has a probability associated with it due to sampling. Hence, it is recommended to run TSNE multiple times & check for results.
